Communicable diseases are leading causes of mortality worldwide. Because infectious diseases have a profound impact on large populations, as experienced with the SARS COV2 pandemic, surveillance, rapid diagnosis and appropriate treatment are paramount to global health. Yet, our understanding of microorganisms is nascent, and conventional pathogen identification techniques are limited.
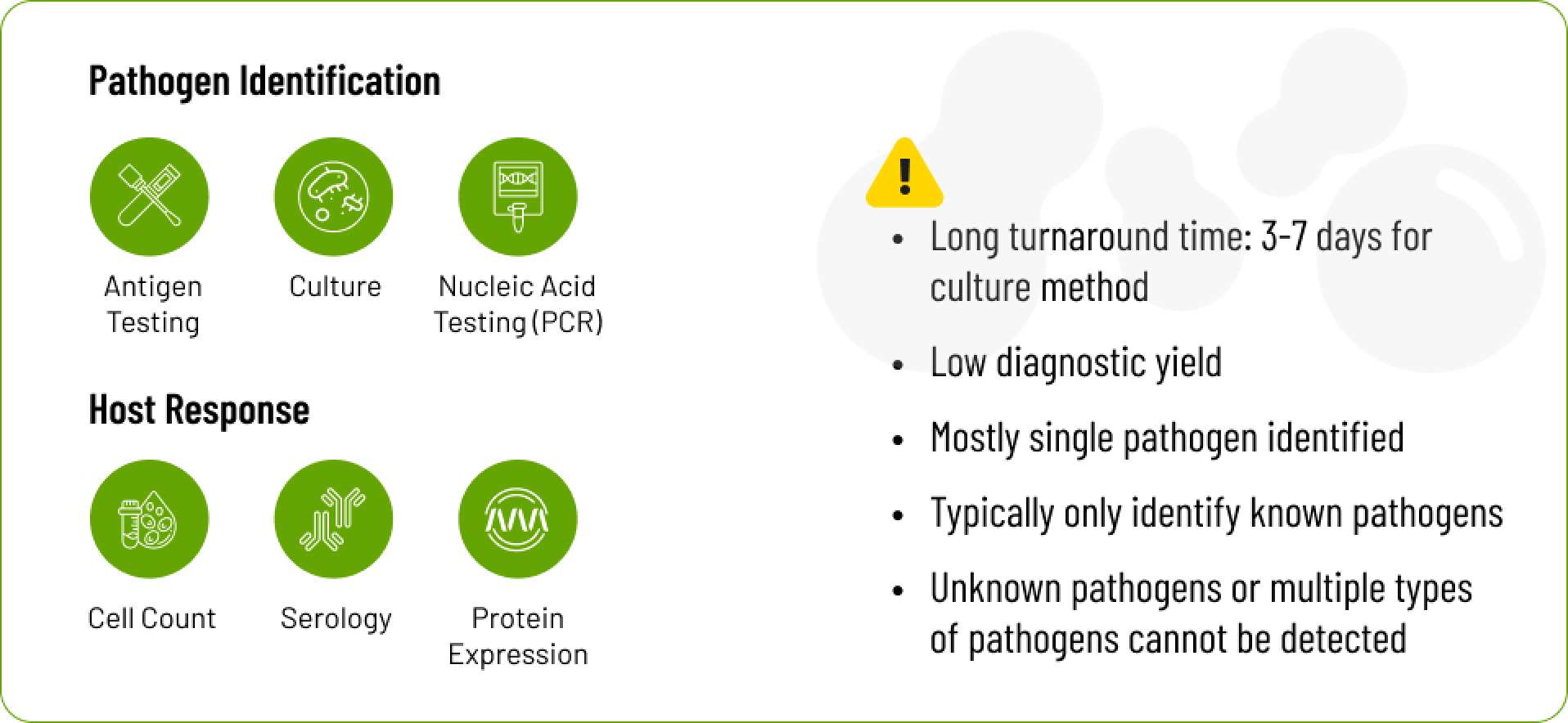
Culturing for bacteria is a century-old method that takes days to weeks and has low diagnostic yield. In an effort to gain more microbial knowledge, molecular modalities, including polymerase chain reaction (PCR) and serology, evolved. Yet, even these advancements are constrained in their ability to detect mixed infections, novel pathogens, and exhibit variable sensitivity and specificity. The absence of a single, comprehensive way to identify pathogens necessitates the use of multiple types of tests, which can lead to delays and escalated healthcare costs.
Metagenomic Next-Generation Sequencing (mNGS) provides an unbiased, culture-independent method to explore microbial dynamics in-depth. Despite the immense potential to accelerate our understanding of microorganisms, mNGS’s slow adoption, especially in clinical settings, is due to technical and cost challenges.
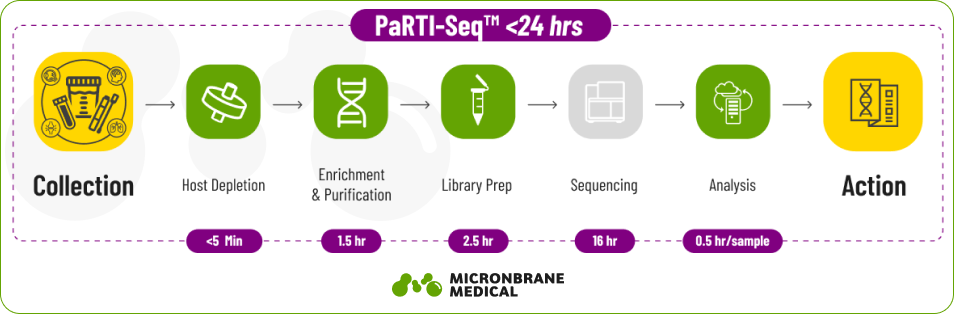
Micronbrane Medical systematically addresses these barriers, with a complete suite of products spanning five categories: specimen collection, host depletion, enrichment and purification, library preparation, and bioinformatic analysis. Our mNGS-enabling technologies together create a fast, efficient and cost-effective assay called PaRTI-Seq to revolutionize infectious disease study, diagnosis, tracking, and treatment.
Overcoming the challenges of mNGS
A primary issue with mNGS is host interference due to the significantly larger size of eukaryotic host genomes compared to prokaryotic genomes. Without any way to remove host cells, only a small proportion of sequencing reads correspond to potential pathogens. The abundance of host genetic material in samples reduces microbiome profile accuracy, leading to negative downstream consequences such as increasing the cost of mNGS and data analysis errors.
However, Micronbrane Medical’s patented Devin™ host depletion filter removes greater than 99 percent of host nucleated cells from up to 10 mL of biological fluid in just five minutes. Devin uses a novel Zwitterionic Interface Ultra-Self-assemble Coating (ZISC) technology not size exclusion to deplete host cells. The Zwitterionic material is a cross-linked polymer with alternating positive and negative ions, which creates a tight hydration layer. The positive and negative ions interact with the hydrophilic proteins of host cells, retaining them in the filter, while allowing microorganisms to pass through unaltered.
By depleting host nucleated cells, microbial DNA can then be enriched. Specifically manufactured with mNGS-grade reagents, the kit minimizes reagent contamination and enhances microbial reads by 10 – 1000 fold, depending on the pathogen load and sample type.
Pre-extraction host depletion necessitates a library kit especially made for low-input DNA, Micronbrane Medical developed the Unison Ultralow Library Preparation Kit. This kit, capable of generating libraries from as little as 10 pg DNA extract, accelerates DNA library preparations through a proprietary tagmentation-based method, condensing fragmentation, adapter ligation, and normalization into a single step.
By effectively reducing host contamination and utilizing the Unison Library Kit, the sequencing depth required for a representative microbiome sample can be significantly decreased, thereby reducing sequencing costs and shortening overall workflow time. Both the nucleic acid purification and NGS library construction steps can be automated for clinical settings.
To streamline the entire mNGS workflow, our PaRTI-Cular Bioinformatic Web App ensures a complete report within 30 minutes per sample. The software automates data quality checks, removal of host genome reads, pathogen identification, and statistical analyses while retaining the data in their own cloud account or a hosted solution.
According to a recent pre-print, the PaRTI-Seq assay streamlined the sample-to-result processes, reducing the overall turn around time and cost per sample. Moreover, the gDNA-based PaRTI-Seq assay performed even better than cell-free DNA-based mNGS, showing an average reads-per-million (RPM) of 2,359 compared to only 95 by a cfDNA-based method. The study concluded that mNGS with the Devin filter was able to recover most of the pathogens identified by clinical BC and achieved the highest diagnostic yield. With the clinical implementation to complete the workflow within 24 hours, it has the potential to overcome slow turnaround and low diagnostic yield issues of traditional microbiology tests.
Utilized as a comprehensive mNGS solution, the combination of PaRTI-Seq and PaRTI-Cular yields cost-effective, actionable results within an unprecedented 24 hour timeframe. By removing the barriers to mNGS use we hope to help expand the use of mNGS in research, clinical, and industrial applications globally.