Infectious diseases have posed significant health challenges throughout human history. While the advent of antibiotics and other antimicrobials was aimed to address this concern, the persistent and widespread use of these agents has led to the emergence of antimicrobial resistance (AMR) and numerous multiple drug-resistant organisms (MDROs).
AMR occurs when bacteria, virus, fungi and parasites change over time and no longer respond to existing treatments. In 2019 the World Health Organization (WHO) described it as one of the top ten global threats to public health – a threat to which science is playing catch-up in its efforts to mitigate.
Close to 5 million lives are lost annually due to drug-resistant infections.2 Projections suggest that without intervention, global deaths attributable to AMR could reach 10 million by 2050 (the same as cancer deaths).3 In addition to high mortality and morbidity, the economic toll of AMR is substantial. In fact, the World Bank estimates that AMR could result in US$ 1 trillion additional healthcare costs by 2050, and US$ 1 – $3.4 trillion gross domestic product (GDP) losses per year by 2030.4
Mechanisms of Antimicrobial Resistance
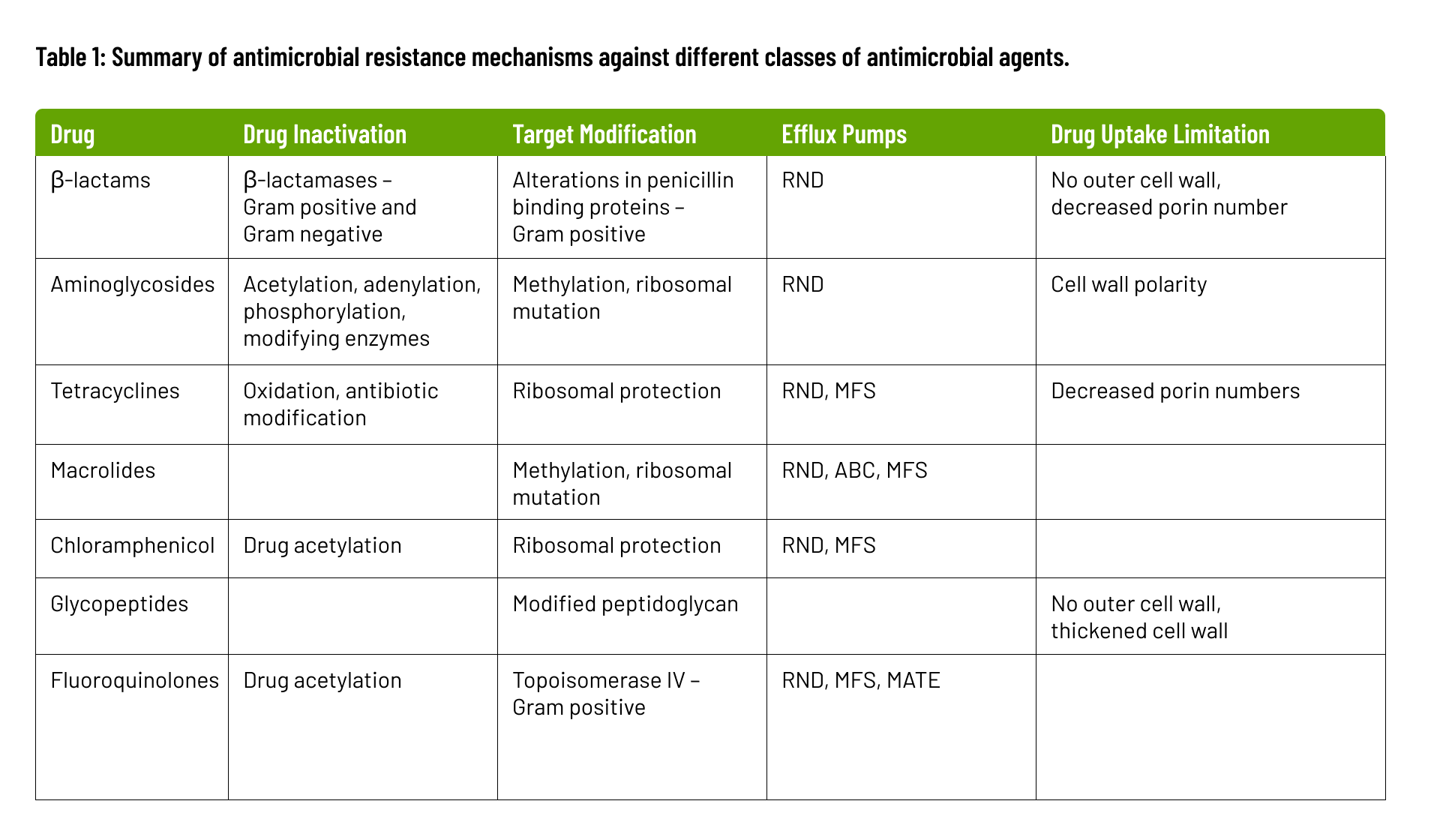
The biochemical and genetic mechanisms that cause AMR, fall into four categories: inactivation of the antimicrobial molecule; target modification; active drug efflux; and drug uptake limitation. These mechanisms are summarized below and in Table 1.
- Inactivation of Antimicrobial Molecules: Microorganisms produce enzymes that deactivate drugs by either destroying them or adding specific chemical components, rendering the antimicrobial ineffective at its target site. These modifying enzymes, catalyzing reactions like acetylation, phosphorylation, and adenylation, induce steric hindrance, diminishing the drug’s affinity for its target and raising the bacterial Minimum Inhibitory Concentration (MIC). β-lactam resistance exemplifies this, employing β-lactamases to break amide bonds in the β-lactam ring, rendering the antimicrobial ineffective. Over 1000 β-lactamases have been identified, with more expected as bacterial evolution continues.
- Target Modification is a key resistance mechanism involving alterations to the antimicrobial target site, impeding proper binding of the antimicrobial molecule. These sites are crucial for cellular functions during antimicrobial action. Mutational changes on the target site can reduce inhibition susceptibility while preserving essential cellular functions. In some cases, inducing resistance through modifying target structures may require additional cellular changes. An example is the acquisition of penicillin-binding transpeptidase (PBP2a) in methicillin-resistant Staphylococcus aureus.
- Active Drug Efflux: Efflux pumps, found in families like the major facilitator superfamily (MFS), small multidrug resistance family (SMR), resistance-nodulation cell division family (RND), ATP-binding cassette family (ABC), and multidrug and toxic compound extrusion family (MATE), have the capacity to expel antimicrobial agents rapidly from the bacterial cell. This expulsion mechanism significantly contributes to multidrug resistance.
- Drug Uptake Limitation: Bacteria vary in their ability to limit drug uptake. The outer membrane composition in organisms like gram-negative bacteria slows antimicrobial penetration. Mycobacteria’s lipid-rich outer membrane hinders hydrophilic drug entry. Organisms lacking a cell wall, such as Mycoplasma, are inherently resistant to cell wall-targeting agents. Biofilm formation protects against immune system attacks and provides defense against antimicrobial agents.
Origins of Antimicrobial Resistance
Microorganisms demonstrate genetic plasticity, enabling them to evolve resistance mechanisms against environmental threats, including antimicrobial agents. The development of antimicrobial resistance involves various processes:
- Mutational Resistance: Susceptible microbial populations undergo mutations in genes affecting drug activity, facilitating cell survival in the presence of antimicrobial agents.
- Spontaneous Mutations: Random mutation events, arising from replication errors or incorrect DNA strand repairs, contribute to antimicrobial resistance.
- Hypermutations: Certain bacterial populations enter transient states of elevated mutation rates under prolonged non-lethal antibiotic pressure. Mutators in this context confer selective advantages.
- Adaptive Mutagenesis: Mutations occur in slowly dividing or non-dividing cells under non-lethal selective pressure, leading to the development of resistant mutants.
- Horizontal Gene Transfer (HGT): Bacterial evolution is influenced by the acquisition of foreign materials through HGT mechanisms, such as transformation, conjugation, and integrons. These processes contribute to the development of antimicrobial resistance.
Addressing Antimicrobial Resistance through Metagenomic Next Generation Sequencing
Metagenomic Next-Generation Sequencing (mNGS) provides unbiased, culture-independent diagnosis and surveillance of resistance mechanisms. Recognized as an indispensable tool, mNGS provides a complete genomic sequence and unparalleled structural detail on individual traits within a population, which contributes to more reliable microbial identification, definitive phylogenetic relationships, and a comprehensive catalog of traits. Additionally, mNGS can also be used for outbreak investigations, microorganism-agnostic infectious disease diagnosis, especially for novel pathogens and appropriate treatment selection.
Understanding the mechanisms and origins of antimicrobial resistance is crucial in developing effective strategies to mitigate its impact. Learn how our Pathogen Real-Time Identification by Sequencing (PaRTI-Seq™) complete mNGS assay provides rapid and accurate identification of resistant pathogens and invaluable insights for the management of AMR. (https://micronbrane.com/products/)
Sources
- Antimicrobial Resistance Collaborators. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet; 399(10325): P629-655. DOI: https://doi.org/10.1016/S0140-6736(21)02724-0
- Dadgostar P. Antimicrobial Resistance: Implications and Costs. Infect Drug Resist. 2019;12:3903-3910 https://doi.org/10.2147/IDR.S234610
- Antimicrobial Resistance Collaborators. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet; 399(10325): P629-655. DOI: https://doi.org/10.1016/S0140-6736(21)02724-0
- The World Bank. 2024 Drug-Resistant Infections: A Threat to Our Economic future. Accessed March 10, 2024 https://www.worldbank.org/en/topic/health/publication/drug-resistant-infections-a-threat-to-our-economic-future
- D’Costa, V. M., McGrann, K. M., Hughes, D. W., and Wright, G. D. Sampling the antibiotic resistome. Science. 2006;311,374–377.
- Jim O’Neill. Tackling drug-resistant infections globally: final report and recommendations the review on antimicrobial resistance chaired. [Online] 2016. Available from: https://amr-review.org/sites/default/files/160518_Final paper_with cover.pdf.
- Founou RC, Founou LL, Essack SY. Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS One. [Online]. 2017;12:e0189621. Available from: https//doi.org/10.1371/journal.pone.0189621
- Shrestha P, Cooper BS, Coast J, et al. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob Resist Infect Control. [Online]. 2018;7(1):98. Available from: doi:10.1186/s13756-018-0384-3
- Martinez JL. General principles of antibiotic resistance in bacteria. Drug Discov Today. 2014;11:33–39.
- Cox G, Wright GD. Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int J Med Microbiol. 2013;303:287–292.
- Davies J. Where have all the antibiotics gone? Can J Infect Dis Med Microbiol. 2006;17:287–90.
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12:35–48.
- G. Spratt. Resistance to antibiotics mediated by target alterations. Science. 1994;264:388–393.
- Bush K. The ABCD’s of β-lactamase nomenclature. [Internet]. 2013;19:549–559. Available from: https://doi.org/10.1007/s10156-013-0640-7
- Bush K, Jacoby GA. Updated functional classification of β-lactamases. Antimicrob Agents Chemother. [Internet]. 2010;54:969–976. Available from: https://doi.org/10.1128/AAC.01009-09
- Warinner, C., Rodrigues, J. F., Vyas, R., Trachsel, C., Shved, N., Grossmann, J., et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 2014;46:336–344.
- J. Piddock, R. Wise. Induction of the SOS response in Escherichia coli by 4-quinolone antimicrobial agents. FEMS Microbiol. Lett. 1987;41:289–294.
- Coculescu BI. Antimicrobial resistance induced by genetic changes. J Med Life. 2009;2:114–123.
- Aminov, R.I. Mackie. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol. Lett. 2007;271:147– 161.
- Angers A., Petrillo M., Patak A., Querci M., Van den Eede G. The role and implementation of next-generation sequencing technologies in the coordinated action plan against antimicrobial resistance. Joint Research Center. 2017;4-7.
- Schmidt, K., Mwaigwisya, S., Crossman, L.C., Doumith, M., Munroe, D., Pires, C., Khan, A.M., Woodford, N., Saunders, N.J., Wain, J., et al. Identification of bacterial pathogens and antimicrobial resistance directly from clinical urines by nanopore-based metagenomic sequencing. J. Antimicrob. Chemother. 2017;72:104–114.
- Köser et al. (2014). Whole-genome sequencing to control antimicrobial resistance. Trends Genet. 30(9):401–7.
- Han et al. (2019). mNGS in clinical microbiology laboratories: on the road to maturity. Crit Rev Microbiol 45(5-6):668-85.
- Balloux et al. (2018), From Theory to Practice: Translating WGS into the Clinic. Trends in Microbiology 26(2).
- Chen et al. (2021). Novel Human Cell Depletion Method For Rapid Pathogen ID by NGS. Labroots Microbiol. Week 10.13140/RG.2.2.23888.64007.
- CDC (2019). Antibiotic Resistance Threats in the US. Atlanta, GA: U.S. HHS Dept, cdc.gov/drugresistance/biggest-threats.html.