Identifying pathogens down to the species level is vital for the proper treatment of diseases. However, many pathogens (like several species of fungi and bacteria) are obligate parasites and cannot be cultured in a lab. The ones that can be grown in a lab are hard to identify compared to higher animals because morphological identification of microbes leads to errors. Medically important parasites are also hard to identify using morphological methods. Nevertheless, over 1 billion people currently suffer from tropical diseases, in most cases caused by a parasite. In the past, it was only possible to identify pathogens and parasites through morphological means. However, recent advances in technology, like Polymerase Chain Reaction (PCR) and high-throughput sequencing, have revolutionized the detection techniques and made molecular diagnostic much more rapid and inexpensive.
DNA barcoding is an approach to identify organisms down to the species level by obtaining short nucleic acid sequences (barcodes) and comparing them with a reference collection of pre-identified species. The technology was inspired by barcodes used in retail products to identify them quickly. DNA barcoding can accurately detect human pathogens and parasites, leading to the proper treatment of diseases. Many PCR-based methods use DNA barcoding to identify human pathogens, especially if the type of pathogen is unknown. In case of a suspected infection, a specific PCR and gel electrophoresis might be sufficient to indicate the presence or absence of the pathogen. Nevertheless, the DNA barcoding method is much more accurate than the PCR-gel electrophoresis method because the latter only detects presence/absence and provides no information about the type of pathogen present if not known earlier.
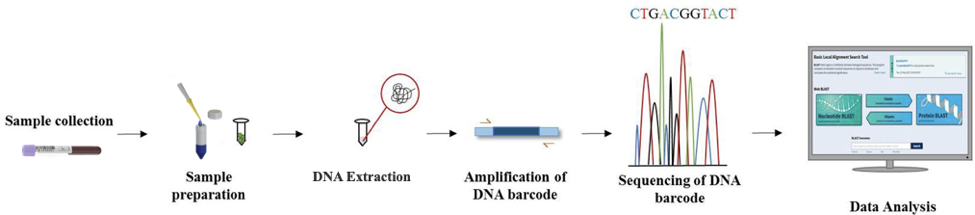
Sequencing a specific DNA segment is required to identify the pathogens through DNA barcoding. The first step to identifying a pathogen is to perform DNA extraction, either from the blood sample or the isolated pathogen. PCR is used to multiply DNA segments from a specific region (barcode in our case). Through Sanger sequencing, a short DNA segment is sequenced. The sequence is searched on a nucleic acid database, such as, NCBI Genbank using the BLAST feature. The database shows the most closely related sequences available in the database and their species. The database also contains information about the sequences, such as the location of sampling, part of the tissue sampled, etc. PCR coupled with Sanger sequencing was the main approach used for DNA barcoding for several years. However, modern technologies like high-throughput next-generation sequencing (NGS) have further improved DNA barcoding. Thanks to the NGS, we can now sequence millions of DNA fragments from thousands of DNA templates in parallel. This helped overcome the limitations of Sanger sequencing and facilitated the generation of DNA barcodes.
Figure 1: Methodology for DNA Barcoding through next-generation sequencing to analyze biodiversity. A similar method can be used to detect human pathogens.
Different molecular markers can be used as barcodes, depending on the type of pathogens or parasites. For a DNA fragment to be considered a barcode, it must meet the following criteria; 1) it should be easy to multiply through PCR. 2) It should have a universal binding site around it so that universal primers can be designed for a collection of organisms. 3) It must be highly conserved within a species and polymorphic between different species. The most used DNA barcode to identify bacteria is the 16s ribosomal RNA gene. Fungi are usually identified through internal transcribed spacer (ITS) sequences. For other eukaryotes, the mitochondrial cytochrome c oxidase subunit I or COI gene is widely used as a DNA barcode marker. Therefore, COI could be the most appropriate marker to detect human parasites or higher animals.
Different fluorescence-related protocols, such as emission wavelength (color), are available for encoding the DNA barcodes. Nucleotide segments can be detected using single-stranded florescent-producing probes. The fluorescence can be detected through a laser and read through a computer to assign a particular species. This method could be useful to develop portable barcode assays to rapidly detect pathogens and help develop barcoded point-of-care bioassays.
Figure 2: Methodology for DNA Barcoding labeled with fluorescence to detect different pathogens causing infectious keratitis.
The first requisite to using DNA barcoding is that the sequence should already be known and present in the databank. If no sequence similar to the pathogen is present in the database, it might mean that there is not enough data available to assign the pathogen to a particular species. Nevertheless, based on similarity, we might know the genus or family of the pathogen. Another limitation of using DNA barcoding is the wrong assignments of sequences in the database. As many databases are open source, people from all over the world can deposit nucleotide arrangements. Some of these sequences might be wrongly assigned a species. Therefore, special caution must be taken to compare the sequence from databases. DNA barcoding is not a replacement for taxonomic and phylogenetic studies. New species still have to be explored and described through both, but DNA barcoding can identify already described and known species. New species can be found through barcoding, but they must go through taxonomic and phylogenetic studies.
Human pathogens are complicated to identify through DNA barcodes. One reason is that most samples contain human DNA, and many markers used to identify pathogens may also present in humans. Therefore, technologies like Devin® blood filtration can be useful to deplete human cells before proceeding with DNA barcoding. Another limitation of DNA barcoding is its accuracy. PaRTI-Seq®, a metagenomic sequencing workflow, can identify potential pathogens from blood samples with 80-90% accuracy within 24 hours. To learn more about PaRTI-Seq® you can watch a short presentational video, download one-pager or e-poster.